Educational Resources for Computational Cell Biology Methods
Video Tutorials

CompCell Biol YouTube - A listing of VCell and COPASI tutorials as well as lecture material from the Annual CCB Online Workshop.
COPASI - A support page with tutorials, user manual, user forum, publications and other technical documentation.
VCell Tutorials - A complete listing of video and pdf format VCell tutorials ranging from simple to complex models.
Webinars & Online Courses

Theory of Living Systems - Theory and computer at the frontier of modern life science.
Biological Modeling - A free course in modeling biological systems at multiple scales.
Coursera - Dynamical Modeling Methods for Systems Biology
Coursera - Introduction to Systems Biology
MITOPENCOURSEWARE - Modeling of Cellular Systems
Publication Resources
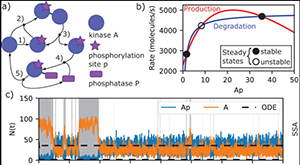
King J, Eroumé KS, Truckenmüller R, Giselbrecht S, Cowan AE, Loew L, Carlier A. Teaching Mathematical Modeling of Cellular Systems with the VCell MathModel. The Biophysicist 2021 Dec.
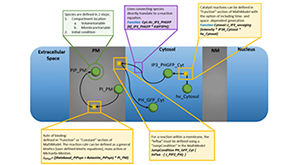
King J, Eroumé KS, Truckenmüller R, Giselbrecht S, Cowan AE, Loew L, Carlier A. Ten steps to investigate a cellular system with mathematical modeling. PLoS Comput Biol. 2021 May;17(5):e1008921.PMCID: PMC8118325.
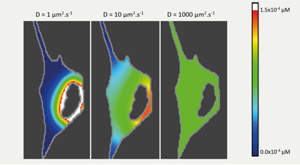
Johnson ME, Chen A, Faeder JR, Henning P, Moraru II, Meier-Schellersheim M, Murphy RF, Prüstel T, Theriot JA, Uhrmacher AM. Quantifying the roles of space and stochasticity in computer simulations for cell biology and cellular biochemistry. Mol Biol Cell. 2021 Jan 15;32(2):186–210. PMID: 33237849.
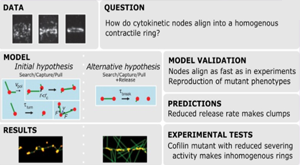
Berro J. Essentially, all models are wrong, but some are useful”—a cross-disciplinary agenda for building useful models in cell biology and biophysics. 2018 Biophys. Rev.. 10 (6), 1637. DOI: 10.1007/s12551-018-0478-4 Brief Bioinform. 2021 Jan 11; PMID: 33423059.
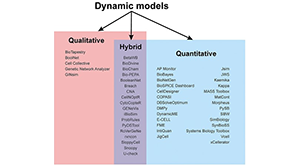
Musilova J, Sedlar K. Tools for time-course simulation in systems biology: a brief overview. Brief Bioinform. 2021 Jan 11; PMID: 33423059.
Tavassoly I, Goldfarb J, Iyengar R. Systems biology primer: the basic methods and approaches. Essays Biochem. 2018 Oct 26;62(4):487–500. PMID: 30287586.
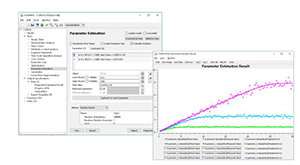
Bergmann FT, Hoops S, Klahn B, Kummer U, Mendes P, Pahle J, Sahle S. COPASI and its applications in biotechnology. J Biotechnol. 2017 Nov 10;261:215–220.PMCID: PMC5623632.
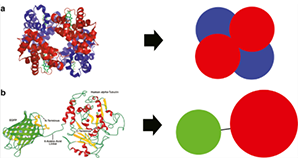
Michalski PJ, Loew LM. SpringSaLaD: A Spatial, Particle-Based Biochemical Simulation Platform with Excluded Volume. Biophys J. 2016 Feb 2;110(3):523–529. PMCID: PMC4744174.
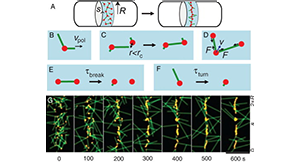
Pollard TD. The value of mechanistic biophysical information for systems-level understanding of complex biological processes such as cytokinesis. Biophys J. 2014 Dec 2;107(11):2499–2507. PMCID: PMC4255220.
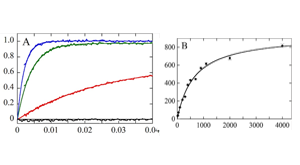
Pollard TD, De La Cruz EM. Take advantage of time in your experiments: a guide to simple, informative kinetics assays. Mol Biol Cell. 2013 Apr;24(8):1103–1110. PMCID: PMC3623632.
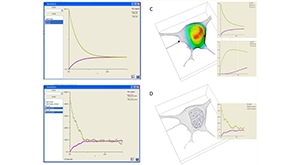
Cowan AE, Moraru II, Schaff JC, Slepchenko BM, Loew LM. Spatial modeling of cell signaling networks. Methods Cell Biol. 2012;110:195–221. PMCID: PMC3519356.
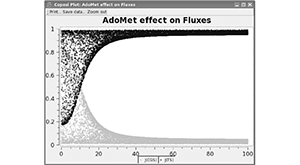
Mendes P, Hoops S, Sahle S, Gauges R, Dada J, Kummer U. Computational modeling of biochemical networks using COPASI. Methods Mol Biol. 2009;500:17–59. PMID: 19399433.