Publications by the Computational Cell Biology Team
Shaikh B, Marupilla G, Wilson M, Blinov ML, Moraru II, Karr JR. RunBioSimulations: an extensible web application that simulates a wide range of computational modeling frameworks, algorithms, and formats. Nucleic Acids Res. 2021 May 21; PMID: 34019658.
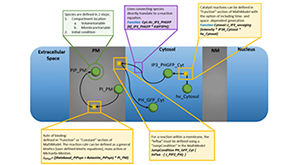
King J, Eroumé KS, Truckenmüller R, Giselbrecht S, Cowan AE, Loew L, Carlier A. Ten steps to investigate a cellular system with mathematical modeling. PLoS Comput Biol. 2021 May;17(5):e1008921. PMCID: PMC8118325.
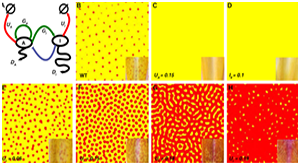
Ding B, Patterson EL, Holalu SV, Li J, Johnson GA, Stanley LE, Greenlee AB, Peng F, Bradshaw HD Jr, Blinov ML, Blackman BK, Yuan YW. Two MYB Proteins in a Self-Organizing Activator-Inhibitor System Produce Spotted Pigmentation Patterns. Curr Biol. 2020 Mar 9;30(5):802-814.e8. PMCID: PMC7156294.
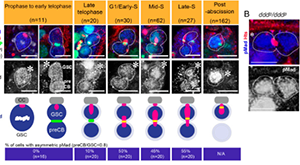
Sardi J, Bener MB, Simao T, Descoteaux AE, Slepchenko BM, Inaba M. Mad dephosphorylation at the nuclear pore is essential for asymmetric stem cell division. Proc Natl Acad Sci U S A. 2021 Mar 30;118(13). PMID: 33753475.
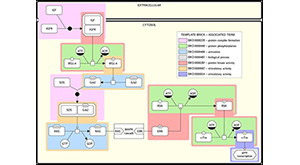
Rougny A, Touré V, Albanese J, Waltemath D, Shirshov D, Sorokin A, Bader GD, Blinov ML, Mazein A. SBGN Bricks Ontology as a tool to describe recurring concepts in molecular networks. Brief Bioinform. 2021 Mar 24; PMID: 33758926.
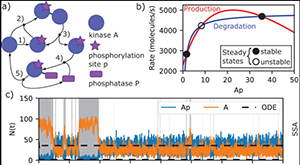
Johnson ME, Chen A, Faeder JR, Henning P, Moraru II, Meier-Schellersheim M, Murphy RF, Prüstel T, Theriot JA, Uhrmacher AM. Quantifying the roles of space and stochasticity in computer simulations for cell biology and cellular biochemistry. Mol Biol Cell. 2021 Jan 15;32(2):186–210. PMID: 33237849.
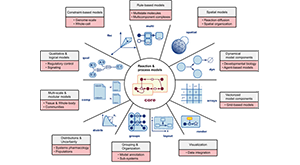
Keating SM et al., SBML Level 3 Community members. SBML Level 3: an extensible format for the exchange and reuse of biological models. Mol Syst Biol. 2020 Aug;16(8):e9110. PMID: 32845085.
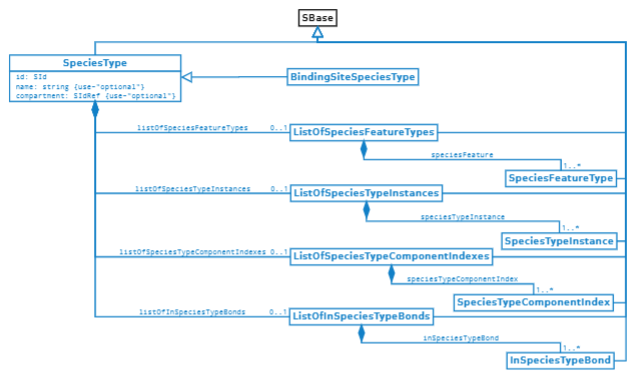
Zhang F, et al., Systems biology markup language (SBML) level 3 package: multistate, multicomponent and multicompartment species, version 1, release 2. J Integr Bioinform. 2020 Jul 6;17(2–3). PMCID: PMC7756619
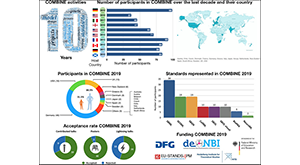
Waltemath D, et al., The first 10 years of the international coordination network for standards in systems and synthetic biology (COMBINE). J Integr Bioinform. 2020 Jun 29;17(2–3). PMCID: PMC7756615.
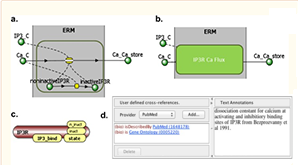
Cowan AE, Mendes P, Blinov ML. ModelBricks-modules for reproducible modeling improving model annotation and provenance. NPJ Syst Biol Appl. 2019 Oct 8;5:37. PMCID: PMC6783478.
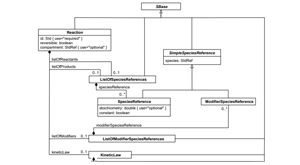
Hucka M, et al., The Systems Biology Markup Language (SBML): Language Specification for Level 3 Version 2 Core Release 2. J Integr Bioinform. 2019 Jun 20;16(2). PMCID: PMC6798823.
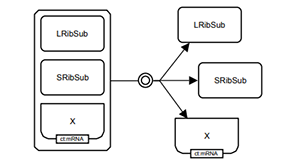
Rougny A, Touré V, Moodie S, Balaur I, Czauderna T, Borlinghaus H, Dogrusoz U, Mazein A, Dräger A, Blinov ML, Villéger A, Haw R, Demir E, Mi H, Sorokin A, Schreiber F, Luna A. Systems Biology Graphical Notation: Process Description language Level 1 Version 2.0. J Integr Bioinform. 2019 Jun 13;16(2). PMCID: PMC6798820.
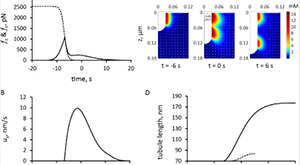
Nickaeen M, Berro J, Pollard TD, Slepchenko BM. Actin assembly produces sufficient forces for endocytosis in yeast. Mol Biol Cell. 2019 22;30(16):2014–2024. PMCID: PMC6727779.
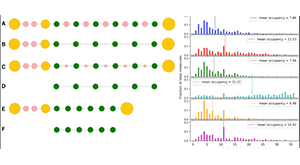
Chattaraj A, Youngstrom M, Loew LM. The Interplay of Structural and Cellular Biophysics Controls Clustering of Multivalent Molecules. Biophys J. 2019 05;116(3):560–572. PMCID: PMC6369576.